Test if a graph has a claw-decomposition
Given two graphs $F$ and $G$, an $F$-decomposition of $G$ is a partition of the edges of $G$ into subgraphs isomorphic to $F$.
First, I am very interested in the case where $F$ is a special subgraph, particularly when F is a claw (i.e. $K_{1,3}$, that is, determining whether a graph has a claw-decomposition. If so, return true and provide a specific decomposition scheme.
For example, the first graph in the following figure has no decomposition (shown for example in M. Hasanvand The existence of planar 4-connected essentially 6-edge-connected graphs with no claw-decompositions. Graphs and Combinatorics, 2023, 39(1): 1. The second graph in the following figure has a claw-decomposition, shown by the four distinct colorings.
I am wondering whether it is possible to write a program to check if a graph has a claw-decomposition. A further question is whether there is a general method for arbitrary subgraphs.
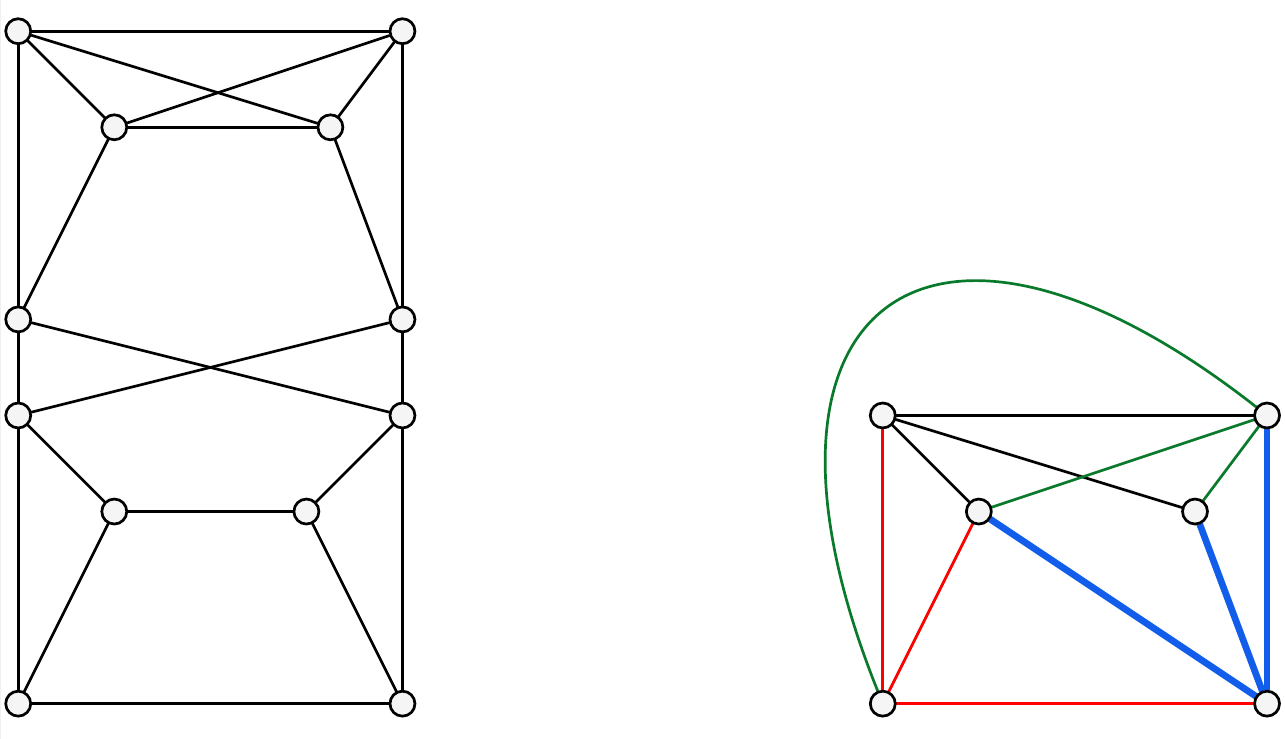
The above two graphs in sage format:
Graph( [(0, 1), (0, 2), (0, 3), (0, 4), (1, 2), (1, 3), (1, 5), (2, 3), (2, 4), (3, 5), (4, 6), (4, 7), (5, 6), (5, 7), (6, 8), (6, 10), (7, 9), (7, 11), (8, 9), (8, 10), (9, 11), (10, 11)])
Graph([(0, 1), (0, 2), (0, 3), (0, 4), (1, 2), (1, 3), (1, 4), (1, 5), (2, 4), (2, 5), (3, 5), (4, 5)])

