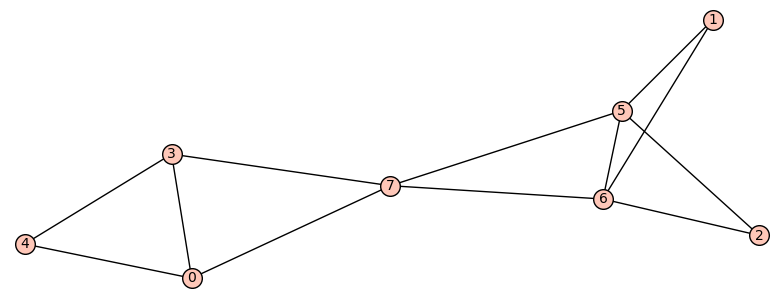
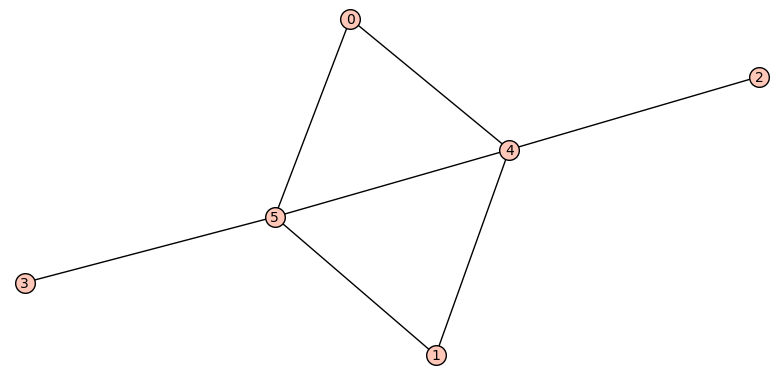
If we only want one pair of graphs for each n, we can store
less data and stop as soon as one pair is found.
With this in mind, we can adapt @tmonteil's code to the following function:
def one_pair(n=3):
r"""
Return a pair of connected graphs on n vertices with same spectral
radius but different diameter and different number of edges.
If no such pair exists, nothing is returned.
"""
from collections import defaultdict
srdm = defaultdict(dict)
for g in graphs.nauty_geng('{} -c'.format(n)):
sr = max(g.adjacency_matrix().eigenvalues())
d = g.diameter()
m = g.num_edges()
if not (d, m) in srdm[sr]:
for (dd, mm) in srdm[sr]:
if d != dd and m != mm:
return sr, (dd, mm), (d, m), srdm[sr][(dd, mm)], g
srdm[sr][(d, m)] = g
It comes as no surprise that the efficiency gain is spectacular.
While running the more exhaustive search for up to 8 vertices
takes on the order of three minutes:
sage: %time pairs = {n: search(n=n) for n in range(3, 9)}
CPU times: user 2min 27s, sys: 4.33 s, total: 2min 32s
Wall time: 3min 10s
with one_pair it takes on the order of one second:
sage: %time pairs = {n: one_pair(n=n) for n in range(3, 9)}
CPU times: user 441 ms, sys: 161 ms, total: 601 ms
Wall time: 1.03 s
and one_pair can go up to 20 vertices in about 30 seconds:
sage: %time pairs = {n: one_pair(n=n) for n in range(3, 21)}
CPU times: user 20.1 s, sys: 1.36 s, total: 21.4 s
Wall time: 25.5 s
To display some concrete examples, introduce a helper function:
sage: from collections import defaultdict
....: def graph_dict(g):
....: d = defaultdict(list)
....: for a, b in g.edges(labels=False):
....: d[a].append(b)
....: return dict(d)
and run a loop over the pairs found by one_pair above, whose output
is ready to be copy-pasted into Sage to explore the examples further.
sage: for n in range(21):
....: if n in pairs and pairs[n]:
....: sr, (da, ma), (db, mb), a, b = pairs[n]
....: sr = AA(sr)
....: sr. exactify()
....: if sr not in QQ:
....: sr = sr.radical_expression()
....: mp = sr.minpoly()
....: print('\nTwo graphs A and B on {} vertices, where'.format(n))
....: print('A has {} edges and diameter {}, '
....: 'B has {} edges and diameter {},'
....: .format(ma, da, mb, db))
....: print('and both have spectral radius r = {} '
....: '(a root of {}):'.format(sr, mp))
....: print('A = Graph({})'.format(graph_dict(a)))
....: print('B = Graph({})'.format(graph_dict(b)))
....:
Two graphs A and B on 6 vertices, where
A has 5 edges and diameter 2, B has 6 edges and diameter 4,
and both have spectral radius r = sqrt(5) (a root of x^2 - 5):
A = Graph({0: [5], 1: [5], 2: [5], 3: [5], 4: [5]})
B = Graph({0: [4, 5], 1: [4, 5], 2: [4], 3: [5]})
Two graphs A and B on 7 vertices, where
A has 6 edges and diameter 2, B has 7 edges and diameter 3,
and both have spectral radius r = sqrt(6) (a root of x^2 - 6):
A = Graph({0: [6], 1: [6], 2: [6], 3: [6], 4: [6], 5: [6]})
B = Graph({0: [5, 6], 1: [5, 6], 2: [6], 3: [6], 4: [6]})
Two graphs A and B on 8 vertices, where
A has 9 edges and diameter 3, B has 10 edges and diameter 4,
and both have spectral radius r = 3 (a root of x - 3):
A = Graph({0: [6, 7], 1: [6, 7], 2: [6], 3: [6], 4: [7], 5: [7], 6: [7]})
B = Graph({0: [6, 7], 1: [6, 7], 2: [6, 7], 3: [6, 7], 4: [6], 5: [7]})
Two graphs A and B on 9 vertices, where
A has 8 edges and diameter 2, B has 10 edges and diameter 4,
and both have spectral radius r = 2*sqrt(2) (a root of x^2 - 8):
A = Graph({0: [8], 1: [8], 2: [8], 3: [8], 4: [8], 5: [8], 6: [8], 7: [8]})
B = Graph({0: [7, 8], 1: [7, 8], 2: [7, 8], 3: [7], 4: [7], 5: [8], 6: [8]})
Two graphs A and B on 10 vertices, where
A has 11 edges and diameter 3, B has 12 edges and diameter 4,
and both have spectral radius r = 3 (a root of x - 3):
A = Graph({0: [7, 8], 1: [8, 9], 2: [8], 3: [9], 4: [9], 5: [9], 6: [9], 7: [8], 8: [9]})
B = Graph({0: [7, 8], 1: [8, 9], 2: [8, 9], 3: [8, 9], 4: [9], 5: [9], 6: [9], 7: [8]})
etc.